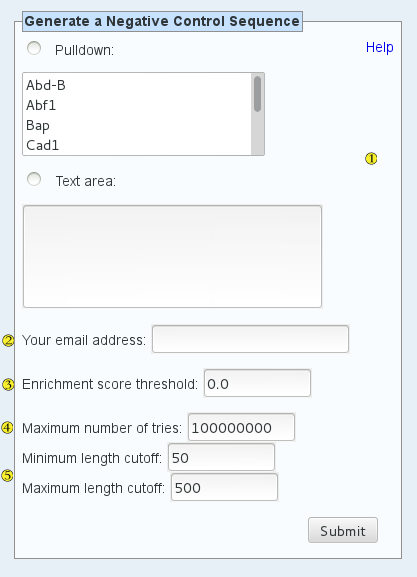

This is a "negative control sequence generator" tool that has been developed in house at the Bulyk lab. You input a list of proteins (and/or specific clones or protein complexes) along with the rest of the parameters listed below, and our algorithm will generate a DNA sequence which has very low probability of any residual binding by any proteins you've selected, based on all the PBM data we've collected in UniPROBE.
The way it works is: we build a file containing all contiguous 8-mers such that every single protein you've selected has scored below a certain enrichment score threshold (which you can also supply) for binding to that 8-mer in every set of PBM data which we've collected for that protein. Then we feed this file into a script which will generate the nonbinding DNA sequence by randomly adding on suitable k-mers to the existing sequence, such that no disallowed 8-mer - i.e., no 8mer not in the input file - will appear at any point in the sequence. Note that since the addition of k-mers is performed randomly, this algorithm is essentially non-deterministic, meaning you will most likely get a very different-looking sequence if you run the tool more than once using the same input proteins and parameter settings.
When it's done, you'll receive an email with the results. Using this tool will always redirect you to the main page, where you can find a summary of the success or failure of your submission at the top of the tool's search box.
 You can input your list of proteins (or clones or complexes) which you use to filter your list of 8mers, either
through the multiple-select pulldown menu at the top or via the text area below.
If you use the text area, enter each protein on its own line, and make sure the names
all match up with names you see in the upper pulldown menu.
You can input your list of proteins (or clones or complexes) which you use to filter your list of 8mers, either
through the multiple-select pulldown menu at the top or via the text area below.
If you use the text area, enter each protein on its own line, and make sure the names
all match up with names you see in the upper pulldown menu.
 The results of this tool will be sent to the email address you provide here.
The results of this tool will be sent to the email address you provide here.
 This is the enrichment score threshold which will be used to select non-binding 8mers.
Only 8mers with enrichment scores below this threshold for every single protein
you selected will be allowed within your negative control sequence.
The lower your threshold, the lower the probability of any residual binding by any
of your proteins, but also the lower the probability that we'll successfully be able
to generate a sequence for you.
This is the enrichment score threshold which will be used to select non-binding 8mers.
Only 8mers with enrichment scores below this threshold for every single protein
you selected will be allowed within your negative control sequence.
The lower your threshold, the lower the probability of any residual binding by any
of your proteins, but also the lower the probability that we'll successfully be able
to generate a sequence for you.
 This parameter represents the maximum number of attempts our algorithm will make to extend
the sequence before it has reached the minimum length cutoff. If it fails to find a suitable
k-mer for extension after this many attempts, it will give up. Increasing this parameter
will increase the likelihood of succesfully generating a sequence, but may
also increase the time necessary to compute your results.
This parameter represents the maximum number of attempts our algorithm will make to extend
the sequence before it has reached the minimum length cutoff. If it fails to find a suitable
k-mer for extension after this many attempts, it will give up. Increasing this parameter
will increase the likelihood of succesfully generating a sequence, but may
also increase the time necessary to compute your results.
 These two cutoffs specify the lower and upper bounds for the length of the sequence
you want to generate. Lowering the minimum cutoff and raising the maximum cutoff
will increase the probability of successful sequence generation. However, you
will probably want to use a more restricted range if you plan on
using these DNA sequences for biological experiments.
These two cutoffs specify the lower and upper bounds for the length of the sequence
you want to generate. Lowering the minimum cutoff and raising the maximum cutoff
will increase the probability of successful sequence generation. However, you
will probably want to use a more restricted range if you plan on
using these DNA sequences for biological experiments.