
This search tool uses the Tomtom program (Gupta S et al., Genome Biol. 2007) from the Meta-MEME suite (v. 3.4) to compare one or more user entered motifs against the Seed and Wobble motifs in the UniPROBE database. The user may enter motifs as frequency matrices, count matrices (.pfm), Meta-MEME 3.x files, or as IUPAC motifs. No more than 30 motifs may be entered at a given time. The output from the Tomtom program, which describes successful motif matches, and logo visualizations that illustrate the motif alignments are then displayed in the Browse page. The most recent versions of these programs can now be downloaded as part of the Meme suite.
Because each run of this tool requires significant processing power from our limited server, we would appreciate it if UniPROBE users refrain from heavy use of this tool. If you wish to run a large number of motifs against our database motifs, we ask that you download the Seed and Wobble motifs of interest from the downloads page and run them on a local version of Tomtom.
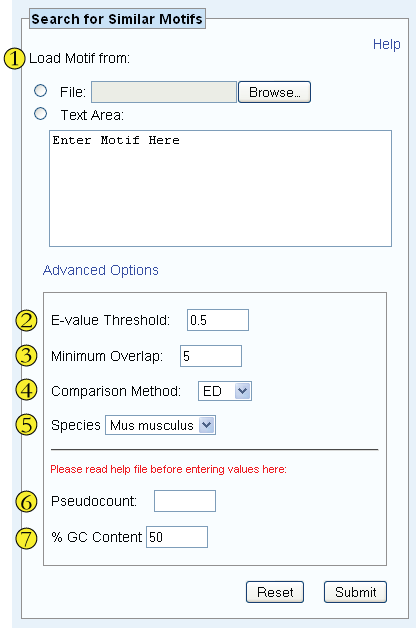
 Load the motifs either by uploading an appropriately formatted file (i.e. contains frequency matrices, count matrices, Meta-MEME 3.x motifs, or IUPAC motifs) or by copying them directly into the Text Area.
Load the motifs either by uploading an appropriately formatted file (i.e. contains frequency matrices, count matrices, Meta-MEME 3.x motifs, or IUPAC motifs) or by copying them directly into the Text Area.
 The Tomtom E-Value "is the expected number of times that a similarity this strong would be observed by chance in a target database of random motifs" (see Tomtom documentation), where values approaching zero indicate a high level of similarity. Only matches scoring below the E-value threshold entered here will be displayed.
The Tomtom E-Value "is the expected number of times that a similarity this strong would be observed by chance in a target database of random motifs" (see Tomtom documentation), where values approaching zero indicate a high level of similarity. Only matches scoring below the E-value threshold entered here will be displayed.
 The minimum number of base pair overlap required for a successful match
The minimum number of base pair overlap required for a successful match
 The algorithm used to quantify the difference between motif patterns. ED stands for Euclidean distance, PCC stands for Pearson correlation coefficient, KLD stands for Kullback- Leiber divergence, and SW stands for Sandelin-Wasserman function.
The algorithm used to quantify the difference between motif patterns. ED stands for Euclidean distance, PCC stands for Pearson correlation coefficient, KLD stands for Kullback- Leiber divergence, and SW stands for Sandelin-Wasserman function.
 Specify the species against which the entered motifs should be tested.
Specify the species against which the entered motifs should be tested.
 The pseudocount value adjusts the motif composition based upon background nucleotide frequency. By default (if the field is left blank), a pseudocount of 1 is used for count matrices, a pseudocount of 0.1 is used for IUPAC motifs, and a pseudocount of 0 is used for Meta-MEME entries and frequency matrices.
The pseudocount value adjusts the motif composition based upon background nucleotide frequency. By default (if the field is left blank), a pseudocount of 1 is used for count matrices, a pseudocount of 0.1 is used for IUPAC motifs, and a pseudocount of 0 is used for Meta-MEME entries and frequency matrices.
 Specify the background GC content as a percentage.
Specify the background GC content as a percentage.