Function get_magic_quotes_gpc() is deprecated
The error 'Function get_magic_quotes_gpc() is deprecated' occurred in the file '' on line 224.
Function get_magic_quotes_gpc() is deprecated
The error 'Function get_magic_quotes_gpc() is deprecated' occurred in the file '' on line 224.
Function get_magic_quotes_gpc() is deprecated
The error 'Function get_magic_quotes_gpc() is deprecated' occurred in the file '' on line 224.
Function get_magic_quotes_gpc() is deprecated
The error 'Function get_magic_quotes_gpc() is deprecated' occurred in the file '' on line 224.
Function get_magic_quotes_gpc() is deprecated
The error 'Function get_magic_quotes_gpc() is deprecated' occurred in the file '' on line 224.
|
|
| -Genomic Data for HAL9 |
Links |
- Protein
- HAL9
|
- UniPROBE Accession Number
- UP00315
|
- Species
- Saccharomyces cerevisiae
|
- Domain
- Zn2Cys6
|
- Uniprot
- Q12180
|
- Synonyms
- Halotolerance protein 9, O0938, YOL089C
|
- IHOP
- 34268
|
- RefSeq
- NP_014552
|
- Description
- Putative transcription factor containing a zinc finger; overexpression increases salt tolerance through increased expression of the ENA1 (Na+/Li+ extrusion pump) gene while gene disruption decrease...
|
- SGD
-
YOL089C
|
| -PBM Motif Data |
- Primary Motif (Seed-and-Wobble)
-
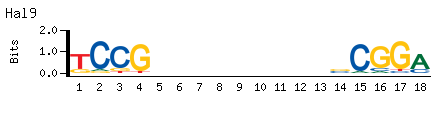
|
- Primary PWM
-
View
Save
|
- Top Kmer
-
TCCG..........CGGA E.S. 0.494
TCCG..........CGGA E.S. 0.494
|
- Primary Motif (BEEML-PBM) (AMADID # 015681)
-
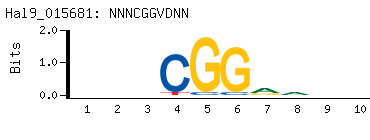
|
- Primary PWM
-
View
|
- Consensus
-
NNNCGGVDNN
NNHBCCGNNN
|
- Downloads
-
Download the
zip
of all Hal9 PBM files
or view the
downloads directory
for individual files.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for Hal9
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID315&sourceDB=uniprobe
|
| -Annotations |
- Target Gene Functional Category Enrichment
-
1) FunSpec analysis run with p<0.005 without Bonferroni correction
(view)
-
2) FunSpec analysis run with p<0.05 with Bonferroni correction
(view)
|
- Conditional Specificity
-
1) Specific conditions (view)
-
2) Condition categories (view)
|
| -HAL9 Datasets |
| Clone ID |
Clone Type |
Last Modified |
Save |
View |
|
y1078
|
DNA binding domain only |
2016-03-24 |

|

|
- Insert sequence
- 1 NVAYPNTIYS PTEHQSQFQT QQNRDISTMM EHTNSNDMSG SGKNLKKRVS
51 KACDHCRKRK IRCDEVDQQT KKCSNCIKFQ LPCTFKHRDE ILKKKRKLEI
101 KHHATPGESL QTSNSISNPV ASSSVPNSGR FELLNGNSPL ESNII
|
|
|
|
Supported Browsers:IE 7.0.6001.18000, Firefox 3.5.11, Google Chrome 5.0.375.125 beta