|
|
| -Genomic Data |
Links |
- Protein
- Lef1
|
- UniPROBE Accession Number
- UP00067
|
- Species
- Mus musculus
|
- Domain
- HMG_box
|
- Uniprot
-
P27782
|
- Name and Synonyms
-
lymphoid enhancer binding factor 1
3000002B05, AI451430, Lef-1, LEF-1, Lymphoid enhancer-binding factor 1, lymphoid enhancer factor 1
|
- IHOP
-
122501
|
- RefSeq
-
NP_034833
|
- Description
- None Available
|
- JASPAR
-
N/A
|
| -PBM Motif Data for Lef1 |
- Primary Motif (Seed-And-Wobble)
-

|
- Primary PWM
-
View
Save
|
- Top Kmer
-
CTTTGATC E.S. 0.498
GATCAAAG E.S. 0.498
|
- Secondary Motif (Seed-And-Wobble)
-

|
- Secondary PWM
-
View
Save
|
- Top Kmer
-
ATCAATCA E.S. 0.476
TGATTGAT E.S. 0.476
|
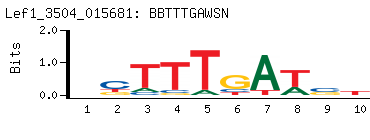
-
Motif (BEEML-PBM) (AMADID # 015681)
(clone pTH3504)
-
|
- PWM
-
View
|
- Consensus
-
BBTTTGAWSN
NSWTCAAAVV
|
-
Motif (BEEML-PBM) (AMADID # 016060) (clone pTH3504)
-
|
- PWM
-
View
|
- Consensus
-
NNHDWNWNNN
NNNWNWHDNN
|
- Downloads
-
Download the
zip
of all Lef1 PBM files
or view the
downloads directory
for individual files.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for Lef1
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID=67&sourceDB=uniprobe
|
| -Lef1 Datasets |
| Clone ID |
Expression |
Array Version |
Expression Protocol |
Clone Type |
Last Modified |
Save |
View |
|
pTH3504 |
#1 |
AMADID #016060 |
E. coli |
DNA binding domain only |
2016-03-24 |

|

|
| Clone ID |
Expression |
Array Version |
Expression Protocol |
Clone Type |
Last Modified |
Save |
View |
|
pTH3504 |
#1 |
AMADID #015681 |
E. coli |
DNA binding domain only |
2016-03-24 |

|

|
- Clone pTH3504 insert sequence
- 1 KPQHEQRKEQ EPKRPHIKKP LNAFMLYMKE MRANVVAECT LKESAAINQI
51 LGRRWHALSR EEQAKYYELA RKERQLHMQL YPGWSARDNY GKKKKRKREK
101 L
- DNA Binding Domains (Pfam) for clone 3504
-
Region 17..85
1 IKKPLNAFML YMKEMRANVV AECTLKESAA INQILGRRWH ALSREEQAKY
51 YELARKERQL HMQLYPGWS
-
|
- References
-
Data are analyzed by array designs with AMADID #s 016060, 015681 in Badis et al., Science 2009
|
|