|
|
| -Genomic Data for Cbf1 |
Links |
- Protein
|
- UniPROBE Accession Number
- UP00397
|
- Species
- Saccharomyces cerevisiae
|
- Domain
- HLH
|
- Swiss-Prot
- P17106
|
- Name and Synonyms
- Name: None Available.
Synonyms:CBP-1, Centromere-binding factor 1, Centromere-binding protein 1, Centromere promoter factor 1, CEP1, CP1, CPF1, J1730, YJR060W
|
- IHOP
- 33772
|
- NCBI RefSeq
- NP_012594
|
- Description
- Helix-loop-helix protein that binds the motif CACRTG, which is present at several sites including MET gene promoters and centromere DNA element I (CDEI); required for nucleosome positioning at this motif; targets Isw1p to DNA
|
- JASPAR
-
None Available
|
| -PBM Motif Data for Cbf1 |
- PBM-Derived DNA Binding Site Motif
(Seed-And-Wobble)
-

|
- PWM
-
Save
|
- Top Kmer
-
ATCACGTG E.S. 0.499
CACGTGAT E.S. 0.499
|
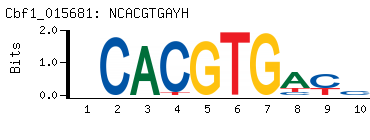
- PBM-Derived DNA Binding Site Motif
(BEEML-PBM) (AMADID # 015681)
-
|
- PWM
-
Save
|
- Consensus
-
NCACGTGAYH
DRTCACGTGN
|
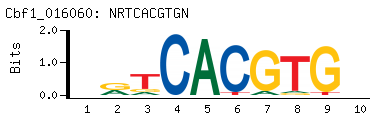
- PBM-Derived DNA Binding Site Motif
(BEEML-PBM) (AMADID # 016060)
-
|
- PWM
-
Save
|
- Consensus
-
NRTCACGTGN
NCACGTGAYN
|
- Downloads
-
Download the
zip
of all Cbf1 PBM files
or view the downloads directory for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for Cbf1
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID=397&sourceDB=uniprobe
|
|