- Top Kmer
-
AAGTACTT (E.S. 0.49877)
- Primary PWM
- View
- Consensus
-
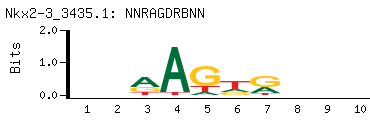
NNRAGDRBNN
- Downloads
- Download the zip of all Nkx2-3 PBM files or view the downloads directory for individual files. Please note that the files which include probe sequence data are protected by an academic research use license, which can be viewed here.
- Link to TFBSshape
-
This link will take you to the corresponding entry for Nkx2-3 in
TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID=190&sourceDB=uniprobe