|
|
| -Genomic Data for CBF1 |
Links |
- Protein
- CBF1
|
- UniPROBE Accession Number
- UP00309
|
- Species
- Saccharomyces cerevisiae
|
- Domain
- HLH
|
- Uniprot
- P17106
|
- Synonyms
- CBP-1, Centromere-binding factor 1, Centromere-binding protein 1, Centromere promoter factor 1, CEP1, CP1, CPF1, J1730, YJR060W
|
- IHOP
- 33772
|
- RefSeq
- NP_012594
|
- Description
- Helix-loop-helix protein that binds the motif CACRTG, which is present at several sites including MET gene promoters and centromere DNA element I (CDEI); required for nucleosome positioning at this motif; targets Isw1p to DNA
|
- SGD
-
YJR060W
|
| -PBM Motif Data |
- Primary Motif (Seed-and-Wobble)
-

|
- Primary PWM
-
View
Save
|
- Top Kmer
-
G.CACGTG.C E.S. 0.498
G.CACGTG.C E.S. 0.498
|
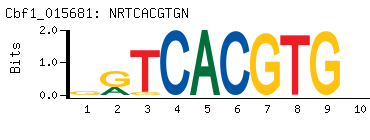
- Primary Motif (BEEML-PBM) (AMADID # 015681)
-
|
- Primary PWM
-
View
|
- Consensus
-
NRTCACGTGN
NCACGTGAYN
|
- Primary Motif (BEEML-PBM) (AMADID # 016060)
-

|
- Primary PWM
-
View
|
- Consensus
-
RRTCACGTGN
NCACGTGAYY
|
- Downloads
-
Download the
zip
of all Cbf1 PBM files
or view the
downloads directory
for individual files.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for Cbf1
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID309&sourceDB=uniprobe
|
| -Annotations |
- Target Gene Functional Category Enrichment
-
1) FunSpec analysis run with p<0.005 without Bonferroni correction
(view)
-
2) FunSpec analysis run with p<0.05 with Bonferroni correction
(view)
|
- Conditional Specificity
-
1) Specific conditions (view)
-
2) Condition categories (view)
|
| -CBF1 Datasets |
| Clone ID |
Clone Type |
Last Modified |
Save |
View |
|
y1001
|
Full length clone |
2014-07-16 |

|

|
- Insert sequence
- 1 MNSLANNNKL STEDEEIHSA RKRGYNEEQN YSEARKKQRD QGLLSQESND
51 GNIDSALLSE GATLKGTQSQ YESGLTSNKD EKGSDDEDAS VAEAAVAATV
101 NYTDLIQGQE DSSDAHTSNQ TNANGEHKDS LNGERAITPS NEGVKPNTSL
151 EGMTSSPMES TQQSKNDTLI PLAEHDRGPE HQQDDEDNDD ADIDLKKDIS
201 MQPGRRGRKP TTLATTDEWK KQRKDSHKEV ERRRRENINT AINVLSDLLP
251 VRESSKAAIL ACAAEYIQKL KETDEANIEK WTLQKLLSEQ NASQLASANE
301 KLQEELGNAY KEIEYMKRVL RKEGIEYEDM HTHKKQENER KSTRSDNPHE
351 A
|
|