|
|
| -Genomic Data |
Links |
- Protein
- Zfp691
|
- UniPROBE Accession Number
- UP00095
|
- Species
- Mus musculus
|
- Domain
- zf-C2H2
|
- Uniprot
-
Q3TDE8
|
- Name and Synonyms
-
Zinc finger protein 691
6430544H17Rik, MGC118413, Zinc finger protein 691, Znf691
|
- IHOP
-
151143
|
- RefSeq
-
NP_898963
|
- Description
- Mus musculus similar to hypothetical protein LOC51058 (LOC242652), mRNA
|
- JASPAR
-
N/A
|
| -PBM Motif Data for Zfp691 |
- Primary Motif (Seed-And-Wobble)
-

|
- Primary PWM
-
View
Save
|
- Top Kmer
-
CAGTGCTC E.S. 0.498
GAGCACTG E.S. 0.498
|
- Secondary Motif (Seed-And-Wobble)
-

|
- Secondary PWM
-
View
Save
|
- Top Kmer
-
AGACTCCT E.S. 0.480
AGGAGTCT E.S. 0.480
|
-
Motif (BEEML-PBM) (AMADID # 015681)
(clone pTH0895)
-
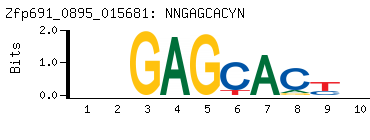
|
- PWM
-
View
|
- Consensus
-
NNGAGCACYN
NRGTGCTCNN
|
-
Motif (BEEML-PBM) (AMADID # 016060) (clone pTH0895)
-
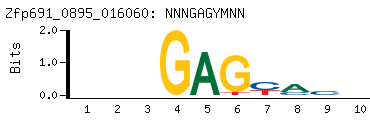
|
- PWM
-
View
|
- Consensus
-
NNNGAGYMNN
NNKRCTCNNN
|
- Downloads
-
Download the
zip
of all Zfp691 PBM files
or view the
downloads directory
for individual files.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for Zfp691
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID=95&sourceDB=uniprobe
|
| -Zfp691 Datasets |
| Clone ID |
Expression |
Array Version |
Expression Protocol |
Clone Type |
Last Modified |
Save |
View |
|
pTH895 |
#1 |
AMADID #016060 |
E. coli |
DNA binding domain only |
2014-08-13 |

|

|
| Clone ID |
Expression |
Array Version |
Expression Protocol |
Clone Type |
Last Modified |
Save |
View |
|
pTH895 |
#1 |
AMADID #015681 |
E. coli |
DNA binding domain only |
2014-08-13 |

|

|
- Clone pTH895 insert sequence
- 1 RDPMLFSSPE TDEKLFICAQ CGKTFNNTSN LRTHQRIHTG EKPYKCSECG
51 KSFSRSSNRI RHERIHLEEK HYQCAKCQES FRRRSDLTTH QQDHLGQRPY
101 RCDICGKSFT QSSTLAVHHR THLEPAPYIC CECGKSFSNS SSFGVHHRTH
151 TGERPYECTE CGRTFSDISN FGAHQRTHRG EKPYRCTLCG KHFSRSSNLI
201 RHQKTHLGEQ DEKDFS
- DNA Binding Domains (Pfam) for clone 895
-
Region 16..38
1 FICAQCGKTF NNTSNLRTHQ RIH
-
-
Region 44..66
1 YKCSECGKSF SRSSNRIRHE RIH
-
Region 72..94
1 YQCAKCQESF RRRSDLTTHQ QDH
-
Region 100..122
1 YRCDICGKSF TQSSTLAVHH RTH
-
Region 128..150
1 YICCECGKSF SNSSSFGVHH RTH
-
Region 156..178
1 YECTECGRTF SDISNFGAHQ RTH
-
Region 184..206
1 YRCTLCGKHF SRSSNLIRHQ KTH
|
- References
-
Data are analyzed by array designs with AMADID #s 016060, 015681 in Badis et al., Science 2009
|
|