- Name and Synonyms
- Name: B-cell lymphoma 6 protein.
Synonyms:B-cell lymphoma 5 protein, B-cell lymphoma 6 protein, BCL5, BCL-5, BCL-6, BCL6A, LAZ3, Protein LAZ-3, ZBTB27, Zinc finger and BTB domain-containing protein 27, Zinc finger protein 51, ZNF51
|
- IHOP
- 86717
|
- Description
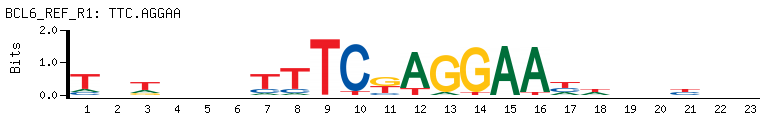
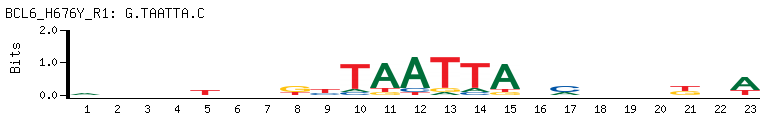
- Transcriptional repressor mainly required for germinal center (GC) formation and antibody affinity maturation which has different mechanisms of action specific to the lineage and biological functions. Forms complexes with different corepressors and histone deacetylases to repress the transcriptional expression of different subsets of target genes. Represses its target genes by binding directly to the DNA sequence 5'-TTCCTAGAA-3' (BCL6-binding site) or indirectly by repressing the transcriptional activity of transcription factors. In GC B-cells, represses genes that function in differentiation, inflammation, apoptosis and cell cycle control, also autoregulates its transcriptional expression and up-regulates, indirectly, the expression of some genes important for GC reactions, such as AICDA, through the repression of microRNAs expression, like miR155. An important function is to allow GC B-cells to proliferate very rapidly in response to T-cell dependent antigens and tolerate the physiological DNA breaks required for immunglobulin class switch recombination and somatic hypermutation without inducing a p53/TP53-dependent apoptotic response. In follicular helper CD4+ T-cells (T(FH) cells), promotes the expression of T(FH)-related genes but inhibits the differentiation of T(H)1, T(H)2 and T(H)17 cells. Also required for the establishment and maintenance of immunological memory for both T- and B-cells. Suppresses macrophage proliferation through competition with STAT5 for STAT-binding motifs binding on certain target genes, such as CCL2 and CCND2. In response to genotoxic stress, controls cell cycle arrest in GC B-cells in both p53/TP53-dependedent and -independent manners. Besides, also controls neurogenesis through the alteration of the composition of NOTCH-dependent transcriptional complexes at selective NOTCH targets, such as HES5, including the recruitment of the deacetylase SIRT1 and resulting in an epigenetic silencing leading to neuronal differentiation.
|
- JASPAR
-
None Available
|