|
|
| -Genomic Data for HLH-2 |
Links |
- Gene
- hlh-2
|
- Species
- $species
|
- Domain
- bHLH
|
- IHOP
-
37860
|
- Swiss-Prot
-
Q17588
|
- UniPROBE Accession Number
- $accession_id
|
- Synonyms
- None Available
|
- $refseq_designator
-
NP_001021581
|
- Wormbase Protein ID
-
WP:CE06191
|
- EdgeDB Promoter
- Phlh-2
|
- Wormbase Gene ID
-
WBGene00001949
|
- Description (from Wormbase)
- hlh-2 encodes a Class I basic helix-loop-helix (bHLH) transcription factor that is the C. elegans ortholog of the mammalian E and Drosophila Daughterless transcriptional activators; HLH-2 activity is required for cell fate specifications occuring during embryonic and larval development that affect such processes as gonadogenesis, male tail formation, and programmed cell death; HLH-2 has been shown to dimerize with at least two C. elegans Acheate-scute homologs, LIN-32, a neural-specific protein with which it functions in male tail development and HLH-3, with which it is coexpressed in the nuclei of embryonic neuronal prescursors and with which it regulates the transcription of the EGL-1 cell death activator in the NSM sister cells; in gonadogenesis, HLH-2 is required for bestowing proAC competence on the cells that undergo the AC/VU (anchor cell/ventral uterine precursor) cell fate decision, for specification, differentiation, and function of the distal tip cell (DTC) and AC, including transcriptional regulation of the LAG-2 Delta-like ligand in the latter, and for formation of the uterine seam cell (utse); genetic analysis also suggests that HLH-2 functions with HLH-14, an additional Acaete-scute homolog, to specify the PVQ/HSN/PHB neuroblast cell lineage; HLH-2 is expressed in all nuclei of early embryos until the ~200-cell stage, when expression becomes increasingly restricted to neuronal cells and their immediate precursors; later expression is detected in, but not limited to, pharyngeal cells, anterior neurons, vulval and uterine muscles, the DTCs, the presumptive and mature AC, the Q neuroblast, and enteric muscles; comparative analysis of transcriptional and translational reporters indicates that hlh-2 is expressed in both the anchor cell and the ventral uterine (VU) precursor, but that expression in the latter is subject to post-transcriptional down-regulation; HLH-2 accumulation in the presumptive AC is the first detectable difference between the AC and VU precursors during the lateral specification event that distinguishes these two cell fates.
|
| -PBM Motif Data for HLH-2 |
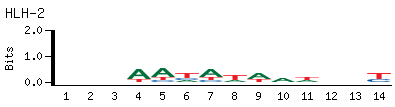
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-
|
- PWM
<dd><a href="./downloads/Cell09/HLH-2/HLH-2.pwm">View</a>   <a href="downfile.php?id=384&gene=HLH-2&folder=Cell09&RC=0">Save</a></dd>
|
- Top Kmer
-
AATATAAT E.S. 0.415
ATTATATT E.S. 0.415
|
- Downloads
-
Download the
zip
of all HLH-2 PBM files
or view the
downloads directory
for individual files.
|
| -PBM Motif Data for HLH-2/CND-1 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
AACA.CTGT E.S. 0.480
ACAG.TGTT E.S. 0.480
|
- Downloads
- Download the zip of all HLH-2/CND-1 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-10 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
CACCTGTT E.S. 0.497
AACAGGTG E.S. 0.497
|
- Downloads
- Download the zip of all HLH-2/HLH-10 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-14 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
ACA.CTGTC E.S. 0.488
GACAG.TGT E.S. 0.488
|
- Downloads
- Download the zip of all HLH-2/HLH-14 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-15 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
CAGCTG.G.C E.S. 0.469
G.C.CAGCTG E.S. 0.469
|
- Downloads
- Download the zip of all HLH-2/HLH-15 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-19 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
ACAGCTGG E.S. 0.478
CCAGCTGT E.S. 0.478
|
- Downloads
- Download the zip of all HLH-2/HLH-19 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-3 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
CCACCTGC E.S. 0.498
GCAGGTGG E.S. 0.498
|
- Downloads
- Download the zip of all HLH-2/HLH-3 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-4 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
A.CACCTGC E.S. 0.499
GCAGGTG.T E.S. 0.499
|
- Downloads
- Download the zip of all HLH-2/HLH-4 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/HLH-8 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
AACAT.TGG E.S. 0.493
CCA.ATGTT E.S. 0.493
|
- Downloads
- Download the zip of all HLH-2/HLH-8 interaction files or view the downloads directory for individual files.
|
| -PBM Motif Data for HLH-2/LIN-32 Heterodimer |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- View
|
- Top Kmer
-
ACAGCTGT E.S. 0.496
ACAGCTGT E.S. 0.496
|
- Downloads
- Download the zip of all HLH-2/LIN-32 interaction files or view the downloads directory for individual files.
|
| -HLH-2 Datasets |
| Gene |
Homodimerizes |
Clone Type |
Last Modified |
Save |
View |
|
hlh-2
|
No |
Full length clone |
2016-03-24 |

|

|
- HLH-2 Insert Sequence
-
1 ADPNSQLTSA TTVATAAIAQ PQVMLPNAYD YPYNIDPTTI QMPDYWSGYH
51 LNPYPPMQTT DIDYSSAFLP THPPTETPAS VAAPTSATSD IKPIHATSST
101 STTAPSTAPA PTSTTDVLEL KPTTAPATNS AETSAIVAPQ PLTNLTAPID
151 AMSSMYTWPQ TYPGYLPPSE DNKASEAVNP YISIPPTYTF GADPSVADFS
201 SYQQQLAGQP NGLGGDTNLV DYNHQFPPAG MSPHFDPNGY PGMTGMPPGS
251 SASSVRNDKS ASRATSRRRV QGPPSSGIPT RHSSSSRLSD NESMSDDKDT
301 DRRSQNNARE RVRVRDINSA FKELGRMCTQ HNQNTERNQT KLGILHNAVS
351 VITQLEEQVR QRNMNPKVMA GMKRKPDDDK MKMLDDNAPS AQFGHPRF
- Mutations away from WormBase HLH-2 wildtype:
- • Methioinine start not included in insert sequence
|
| HLH-2 Interacts With |
Clone Type |
Last Modified |
Save |
View |
| CND-1 |
Full length clone |
2008-07-23 |

|

|
| HLH-10 |
Full length clone |
2008-07-23 |

|

|
| HLH-14 |
Not full length |
2008-07-23 |

|

|
| HLH-15 |
Full length clone |
2008-07-23 |

|

|
| HLH-19 |
Full length clone |
2008-07-23 |

|

|
| HLH-3 |
Not full length |
2008-07-23 |

|

|
| HLH-4 |
Full length clone |
2008-07-23 |

|

|
| HLH-8 |
Full length clone |
2008-07-23 |

|

|
| LIN-32 |
Not full length |
2008-07-23 |

|

|
|