Silencers
A major challenge in biology is to understand how complex gene expression patterns are encoded in the genome. In metazoans, gene expression is regulated in a tissue/cell-type-specific manner predominantly via stretches of noncoding sequence referred to as cis regulatory modules (CRMs). CRMs contain 1 or more DNA binding sites for 1 or more sequence-specific, regulatory transcription factors that function to modulate the expression of target gene(s). CRMs that activate gene expression are typically referred to as enhancers, while those that repress gene expression are referred to as silencers. Transcriptional enhancers activate gene expression in a tissue-specific manner in development and also in adult cells in response to cellular or environmental stimuli. Like enhancers, silencers can function in a cell-type-specific manner. Indeed, silencers may contribute a crucial role in the specification of precise gene expression patterns, thus enabling the establishment of sharp expression domains, such as during development.
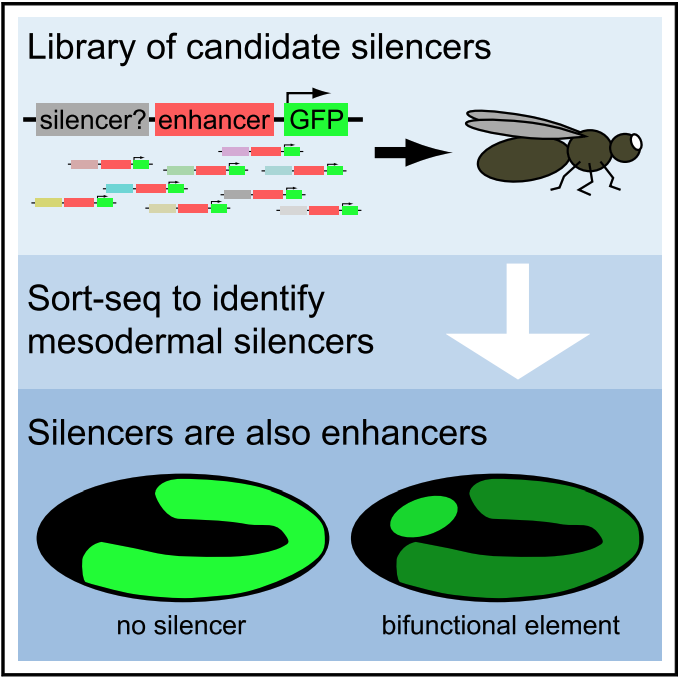
Numerous genomic and computational studies have focused primarily on predicting and characterizing enhancers. In contrast to enhancers, silencers are much less well understood. In a recent study (Gisselbrecht et al., Molecular Cell, 2020), we developed a novel strategy, termed silencer-FACS-Seq (sFS), to screen hundreds of sequences for tissue-specific silencer activity in whole Drosophila embryos. Strikingly, almost all transcriptional silencers we identified were also active enhancers in other cellular contexts. A subset of these silencers form long-range contacts with promoters. Our results challenge the common practice of treating enhancers and silencers as separate classes of regulatory elements and suggest the possibility that thousands or more bifunctional CRMs remain to be discovered in Drosophila and 104 - 105 in human.
We are developing approaches to identify and quantify the activities of tissue-specific silencers, to identify the chromatin signatures of silencers, and to elucidate the regulatory roles of silencer-associated (co-)repressors and DNA sequence motifs. We will focus on the developing embryonic mesoderm in Drosophila melanogaster as our model system. We anticipate that the features and chromatin signatures of silencers identified in this project will be evolutionarily conserved across metazoans, including human.