Structural Biology
I�n� �a�d�d�i�t�i�o�n� �t�o� �d�e�t�e�r�m�i�n�i�n�g� �t�h�e� �D�N�A� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�i�e�s� �o�f� �v�a�r�i�o�u�s� �t�r�a�n�s�c�r�i�p�t�i�o�n� �f�a�c�t�o�r�s� �(�T�F�s�)�,� �w�e� �a�r�e� �a�l�s�o� �i�n�t�e�r�e�s�t�e�d� �i�n� �i�d�e�n�t�i�f�y�i�n�g� �t�h�e� �s�t�r�u�c�t�u�r�a�l� �a�n�d� �m�e�c�h�a�n�i�s�t�i�c� �b�a�s�i�s� �o�f� �t�h�e�s�e� �s�p�e�c�i�f�i�c�i�t�i�e�s�.� �T�F�s� �m�u�s�t� �b�e� �a�b�l�e� �t�o� �b�i�n�d� �t�h�e�i�r� �D�N�A� �t�a�r�g�e�t� �s�i�t�e�s� �w�i�t�h� �h�i�g�h� �a�f�f�i�n�i�t�y� �a�n�d� �d�i�s�t�i�n�g�u�i�s�h� �t�r�u�e� �b�i�n�d�i�n�g� �s�i�t�e�s� �f�r�o�m� �n�o�n�-�t�a�r�g�e�t� �D�N�A�.� �D�e�t�e�r�m�i�n�i�n�g� �t�h�e� �m�e�c�h�a�n�i�s�m�s� �o�f� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�y� �r�e�q�u�i�r�e�s� �s�t�r�u�c�t�u�r�a�l� �s�t�u�d�i�e�s� �o�f� �h�o�w� �t�h�e� �T�F� �r�e�c�o�g�n�i�z�e�s� �D�N�A�.�
T�F�s� �a�r�e� �g�r�o�u�p�e�d� �i�n�t�o� �f�a�m�i�l�i�e�s� �b�a�s�e�d� �o�n� �s�e�q�u�e�n�c�e� �h�o�m�o�l�o�g�y� �w�i�t�h�i�n� �t�h�e� �D�N�A� �b�i�n�d�i�n�g� �d�o�m�a�i�n�,� �a�n�d� �D�N�A� �b�i�n�d�i�n�g� �d�o�m�a�i�n�s� �w�i�t�h�i�n� �a� �f�a�m�i�l�y� �a�d�o�p�t� �s�i�m�i�l�a�r� �t�h�r�e�e�-�d�i�m�e�n�s�i�o�n�a�l� �s�t�r�u�c�t�u�r�e�s�.� �R�e�p�r�e�s�e�n�t�a�t�i�v�e� �s�t�r�u�c�t�u�r�e�s� �o�f� �n�u�m�e�r�o�u�s� �T�F� �f�a�m�i�l�i�e�s� �h�a�v�e� �b�e�e�n� �s�o�l�v�e�d� �i�n� �c�o�m�p�l�e�x� �w�i�t�h� �t�h�e�i�r� �c�o�g�n�a�t�e� �D�N�A� �b�i�n�d�i�n�g� �s�i�t�e� �s�h�o�w�i�n�g� �h�o�w� �e�a�c�h� �f�a�m�i�l�y� �r�e�c�o�g�n�i�z�e�s� �D�N�A�.� �B�e�l�o�w� �a�r�e� �s�o�m�e� �e�x�a�m�p�l�e�s� �o�f� �h�o�w� �w�e� �h�a�v�e� �c�o�m�b�i�n�e�d� �o�u�r� �w�o�r�k� �o�n� �D�N�A� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�i�e�s� �w�i�t�h� �t�h�i�s� �s�t�r�u�c�t�u�r�a�l� �a�p�p�r�o�a�c�h� �i�n� �o�r�d�e�r� �t�o� �u�n�d�e�r�s�t�a�n�d� �m�o�r�e� �t�h�o�r�o�u�g�h�l�y� �t�h�e� �m�e�c�h�a�n�i�s�t�i�c� �b�a�s�i�s� �o�f� �s�p�e�c�i�f�i�c�i�t�y�.� �
�H�o�m�e�o�d�o�m�a�i�n�s�
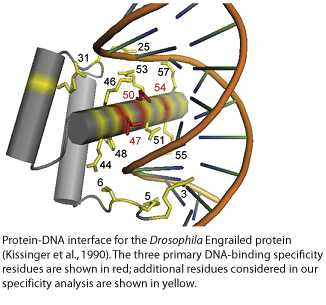
�h�e� �~�6�0� �a�m�i�n�o� �a�c�i�d� �h�o�m�e�o�b�o�x� �d�o�m�a�i�n� �o�r� �`�h�o�m�e�o�d�o�m�a�i�n�'� �i�s� �a� �c�o�n�s�e�r�v�e�d� �D�N�A�-�b�i�n�d�i�n�g� �p�r�o�t�e�i�n� �d�o�m�a�i�n� �b�e�s�t� �k�n�o�w�n� �f�o�r� �i�t�s� �r�o�l�e� �i�n� �t�r�a�n�s�c�r�i�p�t�i�o�n� �r�e�g�u�l�a�t�i�o�n� �d�u�r�i�n�g� �v�e�r�t�e�b�r�a�t�e� �d�e�v�e�l�o�p�m�e�n�t�.� � �T�h�e� �h�o�m�e�o�d�o�m�a�i�n� �b�i�n�d�s� �D�N�A� �p�r�e�d�o�m�i�n�a�n�t�l�y� �t�h�r�o�u�g�h� �i�n�t�e�r�a�c�t�i�o�n�s� �b�e�t�w�e�e�n� �h�e�l�i�x� �3� �(�r�e�c�o�g�n�i�t�i�o�n� �h�e�l�i�x�)� �a�n�d� �t�h�e� �m�a�j�o�r� �g�r�o�o�v�e�.� � �S�i�n�c�e� �m�a�n�y� �h�o�m�e�o�d�o�m�a�i�n�s� �h�a�v�e� �s�i�m�i�l�a�r� �D�N�A� �s�e�q�u�e�n�c�e� �p�r�e�f�e�r�e�n�c�e�s�,� �m�u�c�h� �a�t�t�e�n�t�i�o�n� �h�a�s� �b�e�e�n� �p�a�i�d� �t�o� �t�h�e� �r�o�l�e� �o�f� �p�r�o�t�e�i�n�-�p�r�o�t�e�i�n� �i�n�t�e�r�a�c�t�i�o�n�s� �i�n� �t�a�r�g�e�t� �d�e�f�i�n�i�t�i�o�n�,� �d�e�s�p�i�t�e� �e�v�i�d�e�n�c�e� �t�h�a�t� �t�h�e� �s�e�q�u�e�n�c�e� �s�p�e�c�i�f�i�c�i�t�y� �o�f� �m�o�n�o�m�e�r�s� �c�o�n�t�r�i�b�u�t�e�s� �t�o� �t�a�r�g�e�t�i�n�g� �s�p�e�c�i�f�i�c�i�t�y� �a�n�d� �t�h�a�t� �b�i�n�d�i�n�g� �s�e�q�u�e�n�c�e�s� �d�o� �v�a�r�y�,� �p�a�r�t�i�c�u�l�a�r�l�y� �a�m�o�n�g� �d�i�f�f�e�r�e�n�t� �s�u�b�t�y�p�e�s�.� � �I�t� �h�a�s� �b�e�e�n� �p�r�o�p�o�s�e�d� �t�h�a�t� �t�h�e� �D�N�A� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�y� �o�f� �h�o�m�e�o�d�o�m�a�i�n�s� �i�s� �d�e�t�e�r�m�i�n�e�d� �b�y� �a� �c�o�m�b�i�n�a�t�o�r�i�a�l� �m�o�l�e�c�u�l�a�r� �c�o�d�e� �a�m�o�n�g� �t�h�e� �D�N�A�-�c�o�n�t�a�c�t�i�n�g� �r�e�s�i�d�u�e�s�.
T�h�e� �m�o�u�s�e� �h�o�m�e�o�d�o�m�a�i�n� �c�o�m�p�l�e�m�e�n�t�,� �e�s�t�i�m�a�t�e�d� �a�t� �2�6�0� �d�i�s�t�i�n�c�t� �p�r�o�t�e�i�n�s� �a�n�d� �2�7�5� �i�n�d�i�v�i�d�u�a�l� �h�o�m�e�o�d�o�m�a�i�n�s�,� �i�s� �b�r�o�a�d�l�y� �c�o�n�s�e�r�v�e�d� �a�c�r�o�s�s� �a�n�i�m�a�l�s�.� �W�e� �u�s�e�d� �u�n�i�v�e�r�s�a�l� �p�r�o�t�e�i�n� �b�i�n�d�i�n�g� � �m�i�c�r�o�a�r�r�a�y�s� �(�P�B�M�s�)� �c�o�n�t�a�i�n�i�n�g� �4�1�,�9�4�4� �6�0�-�m�e�r� �p�r�o�b�e�s� �i�n� �w�h�i�c�h� �a�l�l� �p�o�s�s�i�b�l�e� �1�0�-�b�a�s�e� �s�e�q�u�e�n�c�e�s� �a�r�e� �r�e�p�r�e�s�e�n�t�e�d�,� �t�o� �d�e�r�i�v�e� �D�N�A� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�y� �p�r�o�f�i�l�e�s� �f�o�r� �1�6�8� �m�o�u�s�e� �h�o�m�e�o�d�o�m�a�i�n�s�.� � � �O�u�r� �a�n�a�l�y�s�i�s� �s�h�o�w�e�d� �t�h�a�t� �m�o�s�t� �h�o�m�e�o�d�o�m�a�i�n�s� �h�a�v�e� �d�i�s�t�i�n�c�t�i�v�e� �s�e�q�u�e�n�c�e� �p�r�e�f�e�r�e�n�c�e�s�,� �w�h�i�c�h� �m�a�y� �c�o�n�t�r�i�b�u�t�e� �t�o� �t�h�e� �s�t�r�o�n�g� �s�e�l�e�c�t�i�v�e� �p�r�e�s�s�u�r�e� �o�n� �t�h�e�i�r� �a�m�i�n�o� �a�c�i�d� �s�e�q�u�e�n�c�e�s� �a�s� �w�e�l�l� �a�s� � �t�h�e� �b�i�o�l�o�g�i�c�a�l� �s�p�e�c�i�f�i�c�i�t�y� �i�n� �t�a�r�g�e�t� �g�e�n�e�s� �a�n�d� �d�i�v�e�r�s�i�t�y� �i�n� �f�u�n�c�t�i�o�n� �a�m�o�n�g� �t�h�e� �h�o�m�e�o�d�o�m�a�i�n� �p�r�o�t�e�i�n�s�.� � �
I�n� �a�d�d�i�t�i�o�n� �t�o� �b�a�s�e�-�s�p�e�c�i�f�i�c� �c�o�n�t�a�c�t�s� �m�a�d�e� �b�y� �p�o�s�i�t�i�o�n�s� �4�7�,� �5�0�,� �a�n�d� �5�4�,� �w�h�i�c�h� �a�r�e� �b�e�l�i�e�v�e�d� �t�o� �b�e� �t�h�e� �m�a�i�n� �d�e�t�e�r�m�i�n�a�n�t�s� �o�f� �d�i�f�f�e�r�e�n�c�e�s� �i�n� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�y�,� �a�n�d� �r�e�s�i�d�u�e�s� �i�n� �t�h�e� � �N�-�t�e�r�m�i�n�a�l� �a�r�m�,� �w�e� �f�o�u�n�d� �a�d�d�i�t�i�o�n�a�l� �r�e�c�o�g�n�i�t�i�o�n� �p�o�s�i�t�i�o�n�s� �t�h�a�t� �a�r�e� �p�r�e�d�i�c�t�i�v�e� �o�f� �t�h�e� �d�i�f�f�e�r�e�n�c�e�s� �i�n� �D�N�A� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�y� �t�h�a�t� �w�e� �o�b�s�e�r�v�e�d� �f�o�r� �r�e�l�a�t�e�d� �h�o�m�e�o�d�o�m�a�i�n� �p�r�o�t�e�i�n�s�.�
Forkheads
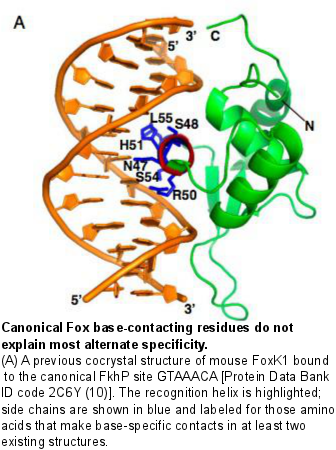
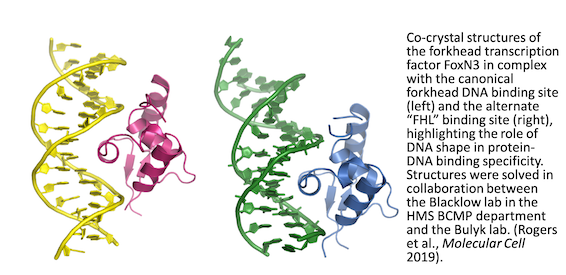
F�o�r�k�h�e�a�d� �T�F�s� �a�r�e� �o�f� �c�e�n�t�r�a�l� �i�m�p�o�r�t�a�n�c�e� �i�n� �d�i�f�f�e�r�e�n�t� �d�e�v�e�l�o�p�m�e�n�t�a�l� �a�n�d� �p�o�s�t�-�n�a�t�a�l� �c�o�n�t�e�x�t�s�,� �i�n�c�l�u�d�i�n�g� �o�r�g�a�n�o�g�e�n�e�s�i�s�,� �e�n�e�r�g�y� �m�e�t�a�b�o�l�i�s�m�,� �h�o�m�e�o�s�t�a�s�i�s�,� �t�h�e� �c�a�r�d�i�o�v�a�s�c�u�l�a�r� �s�y�s�t�e�m�,� �l�i�v�e�r� �r�e�g�e�n�e�r�a�t�i�o�n�,� �h�u�m�a�n� �f�e�m�a�l�e� �r�e�p�r�o�d�u�c�t�i�v�e� �t�i�s�s�u�e�s�,� �i�m�m�u�n�e� �c�e�l�l�s�,� �h�a�i�r� �f�o�l�l�i�c�l�e�s�,� �a�g�i�n�g�,� �a�n�d� �f�o�r� �d�i�s�e�a�s�e�s� �i�n�c�l�u�d�i�n�g� �d�i�a�b�e�t�e�s�,� �c�a�n�c�e�r�,� �g�l�a�u�c�o�m�a�,� �a�n�d� �l�a�n�g�u�a�g�e� �d�i�s�o�r�d�e�r�s�.� �A�s� �d�e�s�c�r�i�b�e�d� �o�n� �o�u�r� �T�r�a�n�s�c�r�i�p�t�i�o�n� �F�a�c�t�o�r� �E�v�o�l�u�t�i�o�n� �w�e�b�p�a�g�e�,� �w�e� �h�a�v�e� �r�e�c�e�n�t�l�y� �i�d�e�n�t�i�f�i�e�d� �m�u�l�t�i�p�l�e� �c�h�a�n�g�e�s� �i�n� �D�N�A� �b�i�n�d�i�n�g� �s�p�e�c�i�f�i�c�i�t�i�e�s� �i�n� �t�h�e� �f�o�r�k�h�e�a�d� �T�F� �f�a�m�i�l�y�.� �C�r�y�s�t�a�l� �s�t�r�u�c�t�u�r�e�s� �o�f� �f�o�r�k�h�e�a�d� �p�r�o�t�e�i�n�s� �i�n� �c�o�m�p�l�e�x� �w�i�t�h� �t�h�e� �c�a�n�o�n�i�c�a�l� �F�K�H� �D�N�A� �b�i�n�d�i�n�g� �s�i�t�e� �m�o�t�i�f� �s�h�o�w� �t�h�a�t� �t�h�e� �f�o�r�k�h�e�a�d� �D�N�A� �b�i�n�d�i�n�g� �d�o�m�a�i�n� �a�d�o�p�t�s� �a� �w�i�n�g�e�d�-�h�e�l�i�x� �f�o�l�d�,� �w�i�t�h� �w�h�i�c�h� �i�t� �p�r�e�s�e�n�t�s� �a� �r�e�c�o�g�n�i�t�i�o�n� �h�e�l�i�x� �t�o� �c�o�n�t�a�c�t� �t�h�e� �m�a�j�o�r� �g�r�o�o�v�e� �o�f� �t�h�e� �D�N�A�.� �T�h�e� �a�m�i�n�o� �a�c�i�d�s� �t�h�a�t� �m�a�k�e� �b�a�s�e�-�s�p�e�c�i�f�i�c� �c�o�n�t�a�c�t�s� �i�n� �t�h�e�s�e� �s�t�r�u�c�t�u�r�e�s� �a�r�e� �w�e�l�l� �c�o�n�s�e�r�v�e�d� �i�n� �t�h�e� �f�a�m�i�l�y�.� �W�e� �f�i�n�d� �t�h�a�t� �a�n� �a�l�t�e�r�n�a�t�e� �s�p�e�c�i�f�i�c�i�t�y� �f�o�r� �t�h�e� �F�H�L� �m�o�t�i�f� �a�r�o�s�e� �t�h�r�e�e� �t�i�m�e�s� �i�n�d�e�p�e�n�d�e�n�t�l�y� �i�n� �t�h�i�s� �f�a�m�i�l�y�.� �

Amino acid positions that contact the bases in the FKH motif are conserved in proteins that recognize the FHL motif. Additionally some forkhead proteins are bispecific: they are capable of binding both the canonical forkhead motif (FKH) as well as the alternate site, the FHL motif. To determine the molecular basis for bispecific DNA recognition, we undertook a combination of biochemical and crystallographic studies of the bispecific forkhead protein FoxN3. In collaboration with Dr. Steve Blacklow’s lab at HMS, we determined co-crystal structures of the human FoxN3 DNA binding domain (DBD) bound to the FKH and FHL sites. Intriguingly, we found that FoxN3 adopts a similar conformation to recognize both sites, using the same amino acids to make contacts with different DNA bases. Strikingly, however, the DNA structure is different in the two complexes. These structures reveal how a single TF binds two unrelated DNA sequences and the importance of DNA shape in the mechanism of bispecific recognition. �