Transcription Factor Evolution
The evolution of gene regulatory networks can involve changes in both cis-regulatory elements (e.g., transcription factor (TF) binding sites) and TFs themselves. Expansion of TF families, through duplication and functional diversification, increases the complexity of gene regulatory networks. Specifically, the divergence of DNA binding specificities within a family of TFs can allow for the gain of novel regulatory functions. While much attention has been paid to the evolution of cis-regulatory sequences, both the mechanisms and effects of changes in TFs are not well understood.
In our study of the forkhead family of TFs (Nakagawa et al., PNAS (2013)), we have found multiple unanticipated changes in DNA binding specificities. While forkhead TF were previously reported to recognize only the canonical FKH motif, we see emergence of binding to a similar alternate motif, the FHL site, multiple times in three independent subfamilies.
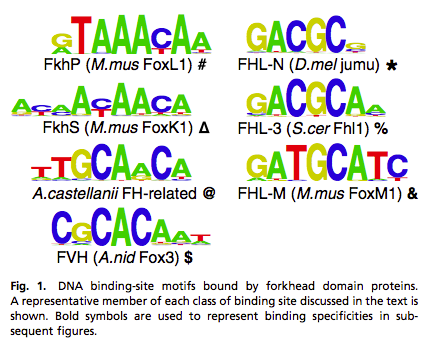
The appearance of similar alternate specificity in three independent forkhead subfamilies raises questions about the mechanism of recognition of the FHL site. For further discussion of such mechanistic questions, please see our Structural Biology webpage.
In the Fox3 subfamily, we see three distinct binding specificities, FKH, FHL, and FVH, among the different yeast species. The functional consequences of these changes on the regulatory circuits are not yet understood. For example, these changes in binding specificities could be accompanied by corresponding changes in cis-regulatory sequence in order to preserve the fundamental regulatory logic, or these specificity changes could allow Fox3 genes in the different species to adopt novel regulatory roles.
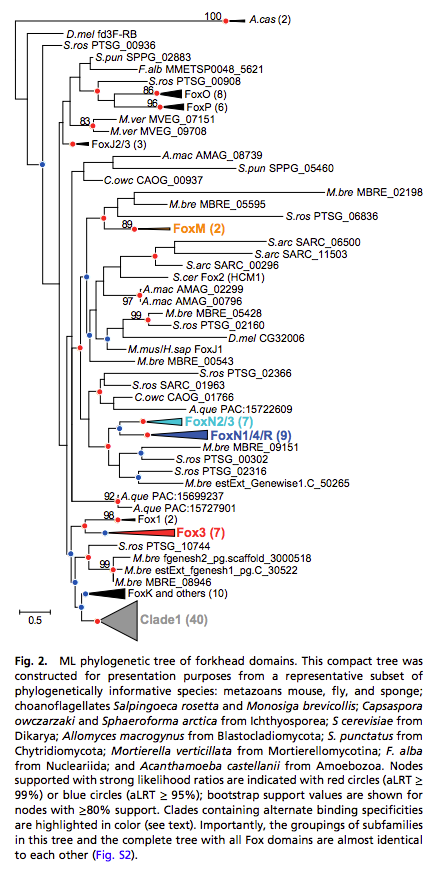
In the metazoan FoxN and FoxM subfamilies, we see proteins that recognize only the FHL motif, as well as proteins that have high specificity for both the FKH and FHL motifs. This bispecificity has not been observed before within a single DNA binding domain. TF bispecificity adds a new dimension to gene regulatory networks, in that these factors can potentially utilize their two motifs to regulate distinct gene sets.
This work on forkhead TFs has shown unanticipated divergence of DNA binding specificities. Subfamilies showing specificity changes provide interesting cases for further investigations of the effects of TF evolution on gene regulatory networks.